
Physics-Informed Deep Learning with Uncertainty Quantification for Weapons Radiography

Project # 23-091 | Year 3 of 3
Arnulfo Gonzaleza, Bence Pappb, Sichen Zhongc, Michael Mortensena
aNevada National Security Sites, bLong Beach State University, cUC Santa Barbara
Abstract
Deep neural network (DNN) models have the potential to provide significant predictive power for a multitude of applications using Nevada National Security Sites (NNSS) image data including radiographs, high-speed footage, accelerator data, and aerial monitoring images. However, for these models to be relevant and assimilated within the Weapons Community, physics-interpretable uncertainty quantification (UQ) of DNN outputs must be provided. This project explores physics-informed DNN models that predict continuous values using image data as inputs, and methods for quantification of the uncertainty associated with the estimates provided by the models. Specifically, we explored fully connected DNNs, Recurrent Neural Networks (RNNs), Convolutional Neural Networks (CNNs), Probabilistic Neural Networks (PNNs), and Bayesian Neural Networks (BNNs). Two methods used for UQ were ensemble models and probabilistic deep learning (PDL) models. Initially, the purpose of this project was to apply Deep Learning (DL) models to radiography data; however, it should be noted, we were unable to acquire the radiography data and instead used public datasets, synthetic data, and reaction history (RH) data..
Background
Signals are extracted from RH film using a semi-automated method. A reader must first manually pick overlay points, baseline points, and inflection points that are subsequently used for Rossi interpolation to complete the signal. This process, however, is extremely time consuming, is subject to human error, and lacks a standardized approach. Two readers can produce significantly different results, affecting the final analysis. Furthermore, there have been numerous failed attempts at automatic signal extraction using image process (and other methods), due to noise, artifacts, and other complex features of RH data. The RH teams at both Lawrence Livermore National Laboratory (LLNL) and Los Alamos National Laboratory (LANL) have long sought a method to automatically perform film reading, particularly given that total number of film ranges in the tens of thousands. We therefore applied DL approaches developed using the Joint Center for Artificial Photosynthesis (JCAP) image dataset and attempted to create models that automatically extract signals from RH film.
Technical Approach
The synthetic timing worm film consisted of 20K samples and were modeled using simple sinusoidal behavior, where the amplitude and frequency are random samples from a Gaussian distribution. The synthetic dataset for yield evolution signal includes 200K samples. RH film signals are taken using the Rossi technique, in which the horizonal oscilloscope sweep is driven sinusoidally in time, while the vertical axis follows the signal amplitude.
The synthetic film, for both datasets, had dimensions 640 x 640 and was down sampled to 64 x 64. The objective was to train numerous DL models (of varying architectures) to predict the signal from film images. However, before training the models, the dimensionality of the film images was reduced from 64 x 64 to 100 x 1 using a variational autoencoder (VAE). VAEs are Bayesian models: they learn the underlying distribution of the data and one can then sample low dimensional representations (latent vectors) from the learned distribution to get reconstructions. We take the latent vectors as the input for our DL models and output the corresponding predicted signals. We only used the full 64 x 64 images for the CNN models. To verify the validity of the latent vectors, we reconstructed the images and found that they have an average structural similarity score of ~0.9. We also found that noise had been significantly reduced in the reconstructed images.
To attempt to improve our DL models, we developed an iterative transfer learning method using the JCAP dataset. That is, instead of training on the entire dataset, we partition the data into classes, train the DL models on an individual class to start, and then iteratively add classes and apply transfer learning. Specifically, we partitioned the JCAP data, consisting of images of metal oxides to predict their corresponding absorption spectra, into classes by number of metals and using clustering. We found that this method was useful for classification using several public image datasets (first learning to pick between two classes, then freezing the original model and retraining it to pick between three, and so on), but it did not offer significant improvement for the regression problem.
.
Results and Technical Accomplishments
We experimented with both probabilistic and non-probabilistic DL models, as well as numerous architectures (RNN, DNN, BNN), and found that the most effective approach was using an ensemble of DNN models with latent space vector representations of input images. This approach was effective when applied to JCAP data, allowing us to both improve on published results (Stein et al.1) and provide uncertainty quantification.
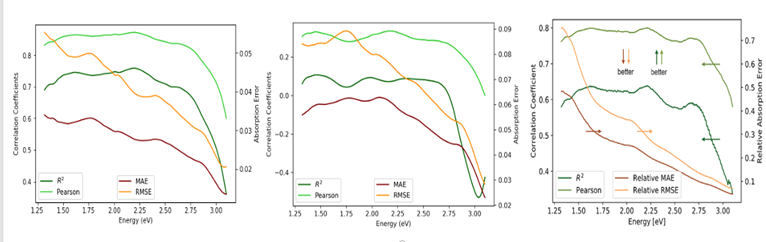
An ensemble of DNNs was extremely effective at extracting timing worm signals from film, giving predictions with an R-squared score ~0.99. However, we found that the Rossi signals cannot be effectively predicted using the DL models we developed and requires further exploration. It should be noted that predicting continuous values using images, particularly for limited or imbalanced datasets, remains an extremely challenging problem with limited literature on the subject. For the JCAP dataset, for example, both our models and the published results produce several predicted spectra with R-squared scores that are approximately zero.
.
Conclusions and Path Forward
The DL models were developed on both LLNL clusters using a virtual environment and Los Alamos Operation’s Athena clusters. User manuals for both clusters were written to help guide future NNSS DL practitioners to efficiently develop models. Mastering a workflow for efficient DL model development, with a supporting infrastructure, marks a significant achievement for this project and the NNSS. Previously, DL model development took place entirely on local machines ill-equipped for handling computationally taxing tasks.
As a result of this project, multiple NNSS staff as well as interns have acquired extensive DL expertise. The team was able to effectively develop DL approaches that can be applied to image datasets to make continuous value predictions with uncertainty quantification. As described in the FY22 final report of this project, our models perform comparably to results published by Stein et al.1 using correlation coefficient and absorption error metrics, with the added benefit of providing uncertainty measures. Moreover, we were able to apply our models to RH film (though with mixed results). Depending on RH funding, these models will likely be incorporated into mission work for further exploration. We will also be applying for future SDRD funding to further develop this work and apply it to radiography data. Currently, LANL and LLNL are using CNN models for radiography image regression without dimensionality reduction or providing uncertainty measures (Proceedings of ASME, 2023), and this work may be beneficial to that effort and the broader DL community.
[1] H. S. Stein et al., Chem. Sci. 10 (2019).
Publications
- Title: pyIFRS Image Processing and Automated Trace Reading Modules
Journal / Conference: 2022 Reaction History Workshop
Year: 2022
Author(s): Arnulfo Gonzalez - Title: Emerging Talent
Journal / Conference: Association for Women in Mathematics
Year: 2022
Author(s): Maggie Lund - Title: LAO Cluster Manual
Journal / Conference: N/A
Year: 2023
Author(s): Bence Papp, Sichen Zhong, Jeremy Fait - Title: Beginners’ Guide to Livermore Computing
Journal / Conference: N/A
Year: 2021
Author(s): Maggie Lund - Title: Supplement to Livermore Computing Guide
Journal / Conference: N/A
Year: 2023
Author(s): Arnulfo Gonzalez
